Electrochemical biosensors

 Electrochemical techniques provide important advantages over optical ones. They are usually less prone to interferences and are better suited for one step target detection directly in blood and other grossly complex samples. Moreover, electrochemical sensors can be easily interrogated with portable instrumentation (see figure) and using disposable, cost-effective printed sensors (screen printed electrodes). We have the possibility to print our own disposable electrodes (graphite, gold etc) using the screen printer machine (DEK245) available at the laboratory of Prof. Palleschi. For all these reasons one of our main goal is to develop novel electrochemical biosensors based on the use of different bio-recognition elements such as DNA, enzymes, peptides etc. and apply them for clinical, food and environmental applications.
Electrochemical techniques provide important advantages over optical ones. They are usually less prone to interferences and are better suited for one step target detection directly in blood and other grossly complex samples. Moreover, electrochemical sensors can be easily interrogated with portable instrumentation (see figure) and using disposable, cost-effective printed sensors (screen printed electrodes). We have the possibility to print our own disposable electrodes (graphite, gold etc) using the screen printer machine (DEK245) available at the laboratory of Prof. Palleschi. For all these reasons one of our main goal is to develop novel electrochemical biosensors based on the use of different bio-recognition elements such as DNA, enzymes, peptides etc. and apply them for clinical, food and environmental applications.
1) Electrochemical-based switches
We propose to immobilize on an electrode surface redox-labelled probes that undergo conformational changes upon target binding. The conformational shift changes the position of the redox label thus resulting in a change of the recorded electrochemical signal. In collaboration with Prof. Plaxco and Prof. Vallée-Bélisle, we have recently demonstrated such sensing principle for the detection of transcription factors (JACS, 2012) and antibodies (JACS, 2012). Press release on this activity.
Check out the short video about this technology!
 We have also recently demonstrated a novel signal-on electrochemical DNA sensor based on the use of a clamp-like DNA probe that binds a complementary target sequence through two distinct and sequential events, which lead to the formation of a triplex DNA structure. We demonstrate that this target-binding mechanism can improve both the affinity and specificity of recognition as opposed to classic probes solely based on Watson–Crick recognition. This work was featured on the cover of Analytical Chemistry (here).
We have also recently demonstrated a novel signal-on electrochemical DNA sensor based on the use of a clamp-like DNA probe that binds a complementary target sequence through two distinct and sequential events, which lead to the formation of a triplex DNA structure. We demonstrate that this target-binding mechanism can improve both the affinity and specificity of recognition as opposed to classic probes solely based on Watson–Crick recognition. This work was featured on the cover of Analytical Chemistry (here).
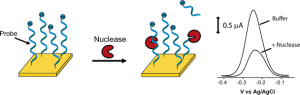
 2) Collisional based E-DNA sensors
2) Collisional based E-DNA sensors
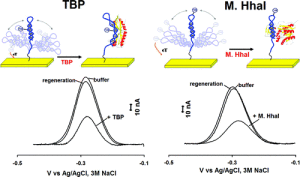
We have developed a reagentless, electrochemical platform for the specific detection of proteins that bind to single- or double-stranded DNA. The sensor is comprised of a double- or singlestranded, redox-tagged DNA probe which is covalently attached to an interrogating electrode. Upon protein binding the current arising from the redox tag is suppressed, indicating the presence of the target. Using this approach we have detected several Transcription Factors at nanomolar concentrations, in minutes, and in a convenient, general, readily reusable, electrochemical format. The approach is specific; we observed no significant cross-reactivity between the sensors. Likewise the approach is selective; it supports, for example, the detection of single strand binding protein directly in crude nuclear extracts.
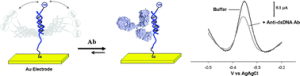 Also, in collaboration with the Plaxco group, we have developed scaffold E-DNA sensors able to detect a range of antibodies including anti-DNA antibodies, diagnostic for auto-immune diseases (JACS, 2009; Chem. Comm, 2010). Media coverage on this activity.
Also, in collaboration with the Plaxco group, we have developed scaffold E-DNA sensors able to detect a range of antibodies including anti-DNA antibodies, diagnostic for auto-immune diseases (JACS, 2009; Chem. Comm, 2010). Media coverage on this activity.
3) Surface chemistry effect on biosensors
 We are studying the role surfaces play in determining biosensor behavior. We are particularly interested in understanding how the surface affects the thermodynamics of a bioreceptor immobilized on it (JACS, 2012). Moreover, we want to shed light on the effect that species absorbed on the surface of an electrode (together with the receptor) have on the signal and general performance of a sensor (Bioelectrochem. 2009). Finally, we are studying the effect that probe density have on the performance of DNA-based biosensors (Langmuir, 2007; manuscript in preparation).
We are studying the role surfaces play in determining biosensor behavior. We are particularly interested in understanding how the surface affects the thermodynamics of a bioreceptor immobilized on it (JACS, 2012). Moreover, we want to shed light on the effect that species absorbed on the surface of an electrode (together with the receptor) have on the signal and general performance of a sensor (Bioelectrochem. 2009). Finally, we are studying the effect that probe density have on the performance of DNA-based biosensors (Langmuir, 2007; manuscript in preparation).
4) Electrochemically-triggered DNA nanomachines
We are studying the possibility to use electrochemistry to trigger several DNA-based switches with future potential as DNA-based nanomachines. More specifically, we want to activate DNA-based switches through the electrochemically controlled release of inputs that can trigger DNA switches. Preliminary results in this context were recently presented during the Biomod competition and we demonstrate the possibility to exogenously trigger a DNA-based nanomachine using a simple electronic input. Because of the simplicity and low-cost associated with basic electronic devices (i.e. battery, power supply, etc) such approach might open the future to a new way to control and trigger DNA-based nanomachines.
