Drug-release nanomachines
Inspired by naturally occurring pH-regulated receptors, in the first work we performed in our lab (you can find the pdf here) in the general field of “drug-release nanomachines” we have proposed a rational approach to introduce pH-induced allostery into a wide range of DNA-based receptors. With this approach we can finely tune the affinity of these model receptors and control the loading/release of their specific target molecule by a simple pH change.
 The regulation of the pH inside or outside the cell and in different tissues of our body represents one of the most efficient strategies that Nature has optimized during evolution to control biological pathways. Nature, for example, uses pH changes to control the load and release of important species. A classic example is represented by haemoglobin, whose affinity towards oxygen gets poorer as the pH decreases. This allows haemoglobin to load oxygen in the lung (where pH is higher) and release it into the muscle tissues (where pH is lower). This pH-induced allostery is often achieved through the exploitation of hydrogen bonds or other pH-dependent interactions in specific domains of the receptor2 that can either activate or inhibit its binding capacity (see figure below).
The regulation of the pH inside or outside the cell and in different tissues of our body represents one of the most efficient strategies that Nature has optimized during evolution to control biological pathways. Nature, for example, uses pH changes to control the load and release of important species. A classic example is represented by haemoglobin, whose affinity towards oxygen gets poorer as the pH decreases. This allows haemoglobin to load oxygen in the lung (where pH is higher) and release it into the muscle tissues (where pH is lower). This pH-induced allostery is often achieved through the exploitation of hydrogen bonds or other pH-dependent interactions in specific domains of the receptor2 that can either activate or inhibit its binding capacity (see figure below).
Inspired by naturally-occurring pH-regulated receptors, we demonstrate here a general strategy to introduce pH-induced allostery into a wide range of DNA-based receptors. More specifically, we re-engineered two different model DNA-based receptors by introducing in their original sequence a pH-dependent domain.
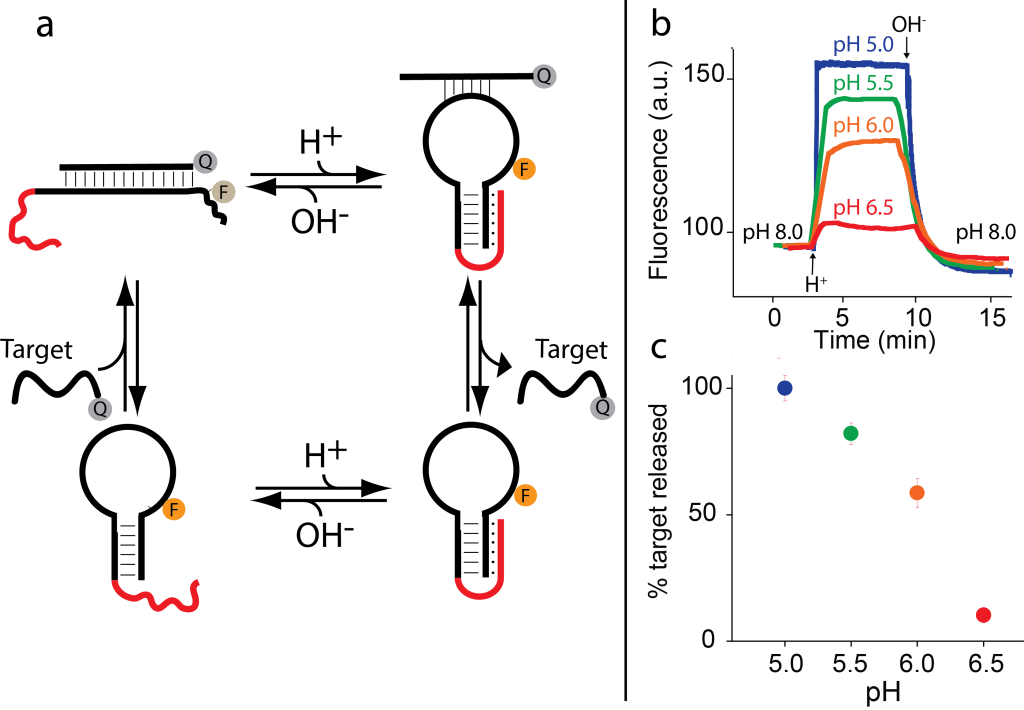 As a first proof-of-principle of this strategy we used the classic stem-loop molecular beacon, an optical labeled DNA-based biosensor. We re-engineer pH-induced allostery in this molecular beacon by designing a pH-dependent triplex-forming stem which is distal from the recognition sequence (loop). Because the additional interactions of the triplex structure will make the stem (and thus the non-binding state) more stable and difficult to be opened by the target, we observe a poorer affinity for the target at acid pHs.
As a first proof-of-principle of this strategy we used the classic stem-loop molecular beacon, an optical labeled DNA-based biosensor. We re-engineer pH-induced allostery in this molecular beacon by designing a pH-dependent triplex-forming stem which is distal from the recognition sequence (loop). Because the additional interactions of the triplex structure will make the stem (and thus the non-binding state) more stable and difficult to be opened by the target, we observe a poorer affinity for the target at acid pHs.
The pH-controlled DNA receptor we have engineered in this work can act as a DNA-based nanomachine that, through pH changes, can reversibly load and release its target in a controlled fashion. By gradually decreasing the pH of the solution from pH 8.0 to pH 6.5, 6.0, 5.5 and 5.0 we can observe a reversible and gradual loading and release of the target from the molecular beacon.
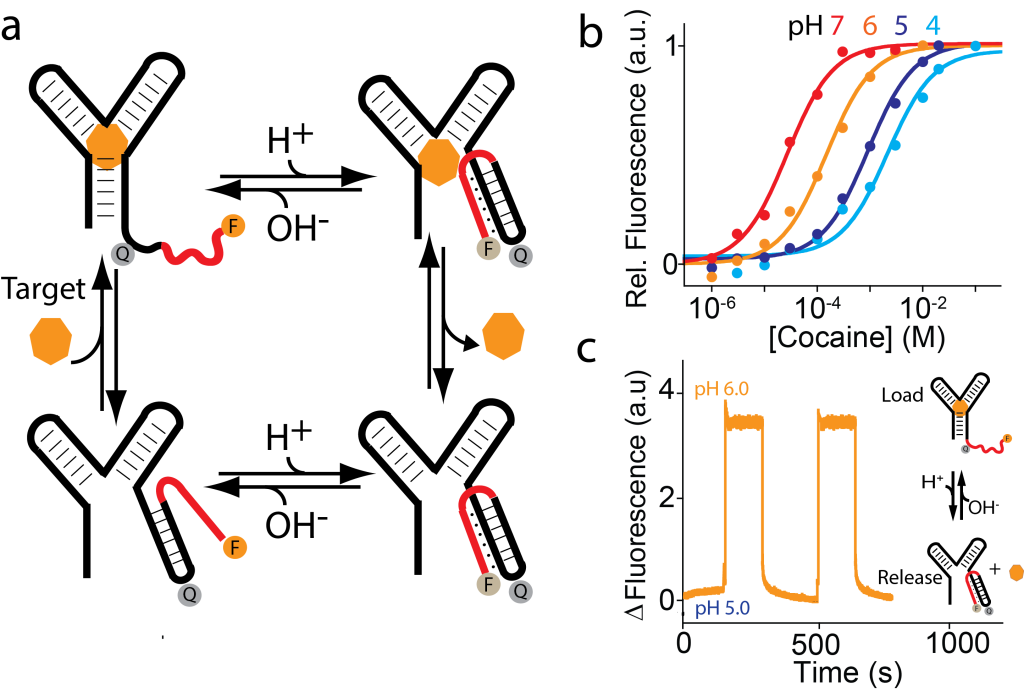 We have also rationally designed pH-induced allostery in a more complex DNA-based aptamer. We have in fact re-engineered the classic cocaine-binding aptamer, which is thought to fold into a three-way junction upon target binding, by introducing at its 3’-end a triplex-DNA forming tail that, when folded, inhibits folding of the aptamer and thus its ability to bind its target. Because triplex formation stabilizes an alternative non-active conformation of the aptamer we can modulate the affinity of this engineered cocaine-binding aptamer by changing the pH of the solution. We also demonstrate the pH-induced load/release of cocaine using this re-engineered aptamer. In the presence of cocaine (i.e. 300 µM) at pH 5.0 no detectable signal increase is observed thus suggesting that no binding occurs. A pH change from 5.0 to 6.0 triggers the aptamer’s ability to bind cocaine and this results in a fluorescence signal increase. Shown are the fluorescence signals subtracted from the background signals at both pH 5.0 and 6.0.
We have also rationally designed pH-induced allostery in a more complex DNA-based aptamer. We have in fact re-engineered the classic cocaine-binding aptamer, which is thought to fold into a three-way junction upon target binding, by introducing at its 3’-end a triplex-DNA forming tail that, when folded, inhibits folding of the aptamer and thus its ability to bind its target. Because triplex formation stabilizes an alternative non-active conformation of the aptamer we can modulate the affinity of this engineered cocaine-binding aptamer by changing the pH of the solution. We also demonstrate the pH-induced load/release of cocaine using this re-engineered aptamer. In the presence of cocaine (i.e. 300 µM) at pH 5.0 no detectable signal increase is observed thus suggesting that no binding occurs. A pH change from 5.0 to 6.0 triggers the aptamer’s ability to bind cocaine and this results in a fluorescence signal increase. Shown are the fluorescence signals subtracted from the background signals at both pH 5.0 and 6.0.
In this first work by our group we have demonstrated a general strategy to re-engineer DNA-based receptors so that their binding affinity can be finely regulated by changes of pH. We have demonstrated the versatility of this strategy by re-engineering two different DNA-based receptors: the classic molecular beacon and the cocaine-binding aptamer. We have rationally inserted, in these two receptors, a pH-sensitive triplex-DNA domain (distal from the recognition site) that upon forming at low pHs, stabilizes an inactive form of the receptor thus reducing its affinity for the target. The strategy proposed here appears suitable to generate DNA-based switching receptors that can be activated/inhibited through pH-changes.